Presentation¶
In [23]:
from IPython.display import HTML
HTML('<img src="images/presentation.png" width="1000">')
Out[23]:
Tools¶
student_depression_dataset¶
In [1]:
import pandas as pd
from matplotlib import pyplot as plt
In [2]:
# Replace 'your_file.csv' with the name or path of your actual CSV file
data = pd.read_csv('~/work/rinchen-khandu/datasets/student_depression_dataset.csv')
data
Out[2]:
| id | Gender | Age | City | Profession | Academic Pressure | Work Pressure | CGPA | Study Satisfaction | Job Satisfaction | Sleep Duration | Dietary Habits | Degree | Have you ever had suicidal thoughts ? | Work/Study Hours | Financial Stress | Family History of Mental Illness | Depression | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | Male | 33.0 | Visakhapatnam | Student | 5.0 | 0.0 | 8.97 | 2.0 | 0.0 | '5-6 hours' | Healthy | B.Pharm | Yes | 3.0 | 1.0 | No | 1 |
| 1 | 8 | Female | 24.0 | Bangalore | Student | 2.0 | 0.0 | 5.90 | 5.0 | 0.0 | '5-6 hours' | Moderate | BSc | No | 3.0 | 2.0 | Yes | 0 |
| 2 | 26 | Male | 31.0 | Srinagar | Student | 3.0 | 0.0 | 7.03 | 5.0 | 0.0 | 'Less than 5 hours' | Healthy | BA | No | 9.0 | 1.0 | Yes | 0 |
| 3 | 30 | Female | 28.0 | Varanasi | Student | 3.0 | 0.0 | 5.59 | 2.0 | 0.0 | '7-8 hours' | Moderate | BCA | Yes | 4.0 | 5.0 | Yes | 1 |
| 4 | 32 | Female | 25.0 | Jaipur | Student | 4.0 | 0.0 | 8.13 | 3.0 | 0.0 | '5-6 hours' | Moderate | M.Tech | Yes | 1.0 | 1.0 | No | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 27896 | 140685 | Female | 27.0 | Surat | Student | 5.0 | 0.0 | 5.75 | 5.0 | 0.0 | '5-6 hours' | Unhealthy | 'Class 12' | Yes | 7.0 | 1.0 | Yes | 0 |
| 27897 | 140686 | Male | 27.0 | Ludhiana | Student | 2.0 | 0.0 | 9.40 | 3.0 | 0.0 | 'Less than 5 hours' | Healthy | MSc | No | 0.0 | 3.0 | Yes | 0 |
| 27898 | 140689 | Male | 31.0 | Faridabad | Student | 3.0 | 0.0 | 6.61 | 4.0 | 0.0 | '5-6 hours' | Unhealthy | MD | No | 12.0 | 2.0 | No | 0 |
| 27899 | 140690 | Female | 18.0 | Ludhiana | Student | 5.0 | 0.0 | 6.88 | 2.0 | 0.0 | 'Less than 5 hours' | Healthy | 'Class 12' | Yes | 10.0 | 5.0 | No | 1 |
| 27900 | 140699 | Male | 27.0 | Patna | Student | 4.0 | 0.0 | 9.24 | 1.0 | 0.0 | 'Less than 5 hours' | Healthy | BCA | Yes | 2.0 | 3.0 | Yes | 1 |
27901 rows × 18 columns
In [3]:
# data cleaning
In [4]:
data.head()
Out[4]:
| id | Gender | Age | City | Profession | Academic Pressure | Work Pressure | CGPA | Study Satisfaction | Job Satisfaction | Sleep Duration | Dietary Habits | Degree | Have you ever had suicidal thoughts ? | Work/Study Hours | Financial Stress | Family History of Mental Illness | Depression | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | Male | 33.0 | Visakhapatnam | Student | 5.0 | 0.0 | 8.97 | 2.0 | 0.0 | '5-6 hours' | Healthy | B.Pharm | Yes | 3.0 | 1.0 | No | 1 |
| 1 | 8 | Female | 24.0 | Bangalore | Student | 2.0 | 0.0 | 5.90 | 5.0 | 0.0 | '5-6 hours' | Moderate | BSc | No | 3.0 | 2.0 | Yes | 0 |
| 2 | 26 | Male | 31.0 | Srinagar | Student | 3.0 | 0.0 | 7.03 | 5.0 | 0.0 | 'Less than 5 hours' | Healthy | BA | No | 9.0 | 1.0 | Yes | 0 |
| 3 | 30 | Female | 28.0 | Varanasi | Student | 3.0 | 0.0 | 5.59 | 2.0 | 0.0 | '7-8 hours' | Moderate | BCA | Yes | 4.0 | 5.0 | Yes | 1 |
| 4 | 32 | Female | 25.0 | Jaipur | Student | 4.0 | 0.0 | 8.13 | 3.0 | 0.0 | '5-6 hours' | Moderate | M.Tech | Yes | 1.0 | 1.0 | No | 0 |
In [6]:
data.columns
Out[6]:
Index(['id', 'Gender', 'Age', 'City', 'Profession', 'Academic Pressure',
'Work Pressure', 'CGPA', 'Study Satisfaction', 'Job Satisfaction',
'Sleep Duration', 'Dietary Habits', 'Degree',
'Have you ever had suicidal thoughts ?', 'Work/Study Hours',
'Financial Stress', 'Family History of Mental Illness', 'Depression'],
dtype='object')
In [7]:
data.describe()
Out[7]:
| id | Age | Academic Pressure | Work Pressure | CGPA | Study Satisfaction | Job Satisfaction | Work/Study Hours | Depression | |
|---|---|---|---|---|---|---|---|---|---|
| count | 27901.000000 | 27901.000000 | 27901.000000 | 27901.000000 | 27901.000000 | 27901.000000 | 27901.000000 | 27901.000000 | 27901.000000 |
| mean | 70442.149421 | 25.822300 | 3.141214 | 0.000430 | 7.656104 | 2.943837 | 0.000681 | 7.156984 | 0.585499 |
| std | 40641.175216 | 4.905687 | 1.381465 | 0.043992 | 1.470707 | 1.361148 | 0.044394 | 3.707642 | 0.492645 |
| min | 2.000000 | 18.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 35039.000000 | 21.000000 | 2.000000 | 0.000000 | 6.290000 | 2.000000 | 0.000000 | 4.000000 | 0.000000 |
| 50% | 70684.000000 | 25.000000 | 3.000000 | 0.000000 | 7.770000 | 3.000000 | 0.000000 | 8.000000 | 1.000000 |
| 75% | 105818.000000 | 30.000000 | 4.000000 | 0.000000 | 8.920000 | 4.000000 | 0.000000 | 10.000000 | 1.000000 |
| max | 140699.000000 | 59.000000 | 5.000000 | 5.000000 | 10.000000 | 5.000000 | 4.000000 | 12.000000 | 1.000000 |
In [8]:
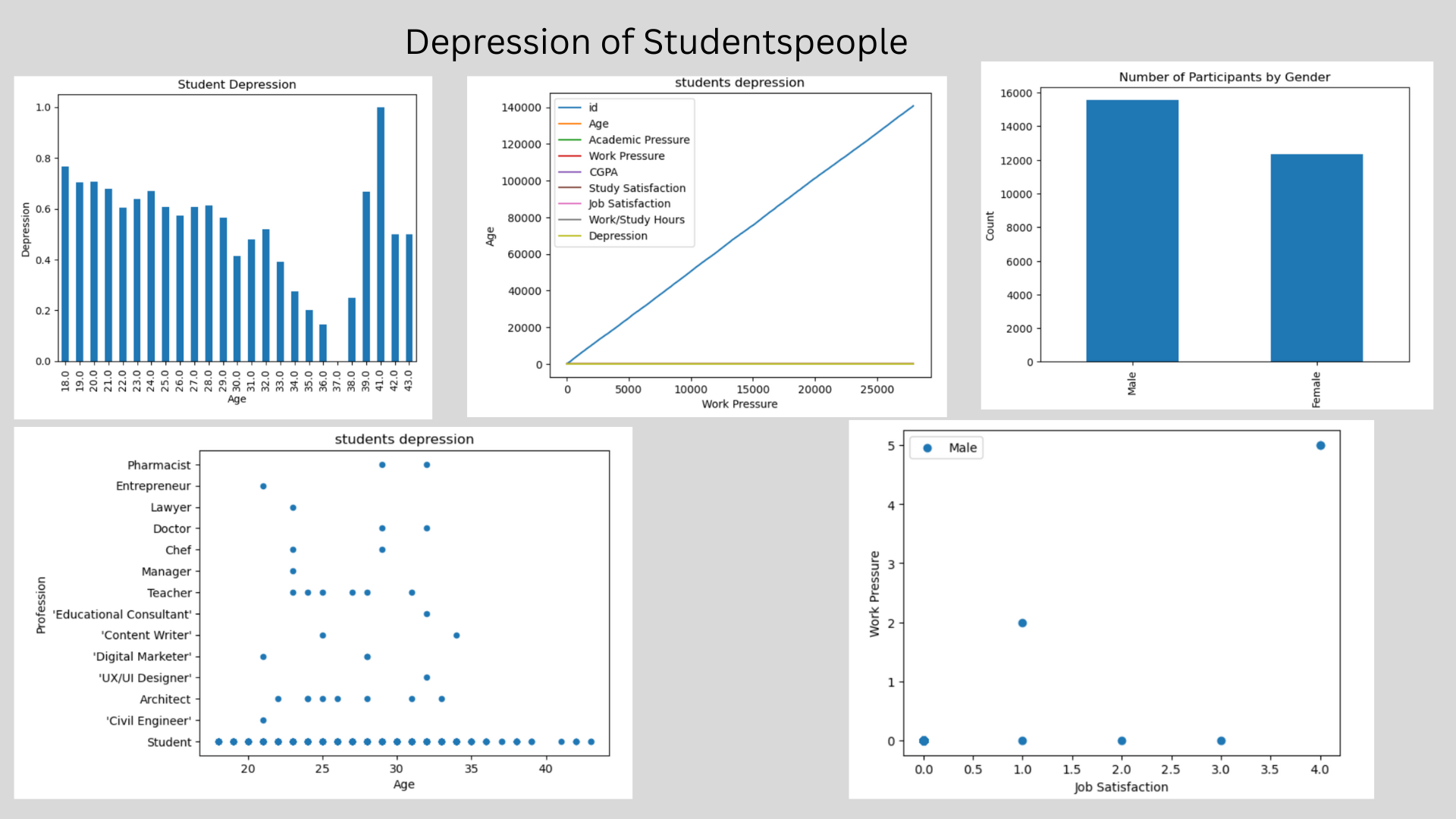
data["Gender"].value_counts()
Out[8]:
Gender Male 15547 Female 12354 Name: count, dtype: int64
In [9]:
data.tail()
Out[9]:
| id | Gender | Age | City | Profession | Academic Pressure | Work Pressure | CGPA | Study Satisfaction | Job Satisfaction | Sleep Duration | Dietary Habits | Degree | Have you ever had suicidal thoughts ? | Work/Study Hours | Financial Stress | Family History of Mental Illness | Depression | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27896 | 140685 | Female | 27.0 | Surat | Student | 5.0 | 0.0 | 5.75 | 5.0 | 0.0 | '5-6 hours' | Unhealthy | 'Class 12' | Yes | 7.0 | 1.0 | Yes | 0 |
| 27897 | 140686 | Male | 27.0 | Ludhiana | Student | 2.0 | 0.0 | 9.40 | 3.0 | 0.0 | 'Less than 5 hours' | Healthy | MSc | No | 0.0 | 3.0 | Yes | 0 |
| 27898 | 140689 | Male | 31.0 | Faridabad | Student | 3.0 | 0.0 | 6.61 | 4.0 | 0.0 | '5-6 hours' | Unhealthy | MD | No | 12.0 | 2.0 | No | 0 |
| 27899 | 140690 | Female | 18.0 | Ludhiana | Student | 5.0 | 0.0 | 6.88 | 2.0 | 0.0 | 'Less than 5 hours' | Healthy | 'Class 12' | Yes | 10.0 | 5.0 | No | 1 |
| 27900 | 140699 | Male | 27.0 | Patna | Student | 4.0 | 0.0 | 9.24 | 1.0 | 0.0 | 'Less than 5 hours' | Healthy | BCA | Yes | 2.0 | 3.0 | Yes | 1 |
Data Visualization¶
In [10]:
data.plot(kind = "line", title = "students depression", xlabel = "Work Pressure", ylabel = "Age")
data
Out[10]:
| id | Gender | Age | City | Profession | Academic Pressure | Work Pressure | CGPA | Study Satisfaction | Job Satisfaction | Sleep Duration | Dietary Habits | Degree | Have you ever had suicidal thoughts ? | Work/Study Hours | Financial Stress | Family History of Mental Illness | Depression | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | Male | 33.0 | Visakhapatnam | Student | 5.0 | 0.0 | 8.97 | 2.0 | 0.0 | '5-6 hours' | Healthy | B.Pharm | Yes | 3.0 | 1.0 | No | 1 |
| 1 | 8 | Female | 24.0 | Bangalore | Student | 2.0 | 0.0 | 5.90 | 5.0 | 0.0 | '5-6 hours' | Moderate | BSc | No | 3.0 | 2.0 | Yes | 0 |
| 2 | 26 | Male | 31.0 | Srinagar | Student | 3.0 | 0.0 | 7.03 | 5.0 | 0.0 | 'Less than 5 hours' | Healthy | BA | No | 9.0 | 1.0 | Yes | 0 |
| 3 | 30 | Female | 28.0 | Varanasi | Student | 3.0 | 0.0 | 5.59 | 2.0 | 0.0 | '7-8 hours' | Moderate | BCA | Yes | 4.0 | 5.0 | Yes | 1 |
| 4 | 32 | Female | 25.0 | Jaipur | Student | 4.0 | 0.0 | 8.13 | 3.0 | 0.0 | '5-6 hours' | Moderate | M.Tech | Yes | 1.0 | 1.0 | No | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 27896 | 140685 | Female | 27.0 | Surat | Student | 5.0 | 0.0 | 5.75 | 5.0 | 0.0 | '5-6 hours' | Unhealthy | 'Class 12' | Yes | 7.0 | 1.0 | Yes | 0 |
| 27897 | 140686 | Male | 27.0 | Ludhiana | Student | 2.0 | 0.0 | 9.40 | 3.0 | 0.0 | 'Less than 5 hours' | Healthy | MSc | No | 0.0 | 3.0 | Yes | 0 |
| 27898 | 140689 | Male | 31.0 | Faridabad | Student | 3.0 | 0.0 | 6.61 | 4.0 | 0.0 | '5-6 hours' | Unhealthy | MD | No | 12.0 | 2.0 | No | 0 |
| 27899 | 140690 | Female | 18.0 | Ludhiana | Student | 5.0 | 0.0 | 6.88 | 2.0 | 0.0 | 'Less than 5 hours' | Healthy | 'Class 12' | Yes | 10.0 | 5.0 | No | 1 |
| 27900 | 140699 | Male | 27.0 | Patna | Student | 4.0 | 0.0 | 9.24 | 1.0 | 0.0 | 'Less than 5 hours' | Healthy | BCA | Yes | 2.0 | 3.0 | Yes | 1 |
27901 rows × 18 columns
Scatter plot¶
In [11]:
data.plot(kind = "scatter", title = "students depression", x = "Age", y = "Profession")
data
Out[11]:
| id | Gender | Age | City | Profession | Academic Pressure | Work Pressure | CGPA | Study Satisfaction | Job Satisfaction | Sleep Duration | Dietary Habits | Degree | Have you ever had suicidal thoughts ? | Work/Study Hours | Financial Stress | Family History of Mental Illness | Depression | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | Male | 33.0 | Visakhapatnam | Student | 5.0 | 0.0 | 8.97 | 2.0 | 0.0 | '5-6 hours' | Healthy | B.Pharm | Yes | 3.0 | 1.0 | No | 1 |
| 1 | 8 | Female | 24.0 | Bangalore | Student | 2.0 | 0.0 | 5.90 | 5.0 | 0.0 | '5-6 hours' | Moderate | BSc | No | 3.0 | 2.0 | Yes | 0 |
| 2 | 26 | Male | 31.0 | Srinagar | Student | 3.0 | 0.0 | 7.03 | 5.0 | 0.0 | 'Less than 5 hours' | Healthy | BA | No | 9.0 | 1.0 | Yes | 0 |
| 3 | 30 | Female | 28.0 | Varanasi | Student | 3.0 | 0.0 | 5.59 | 2.0 | 0.0 | '7-8 hours' | Moderate | BCA | Yes | 4.0 | 5.0 | Yes | 1 |
| 4 | 32 | Female | 25.0 | Jaipur | Student | 4.0 | 0.0 | 8.13 | 3.0 | 0.0 | '5-6 hours' | Moderate | M.Tech | Yes | 1.0 | 1.0 | No | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 27896 | 140685 | Female | 27.0 | Surat | Student | 5.0 | 0.0 | 5.75 | 5.0 | 0.0 | '5-6 hours' | Unhealthy | 'Class 12' | Yes | 7.0 | 1.0 | Yes | 0 |
| 27897 | 140686 | Male | 27.0 | Ludhiana | Student | 2.0 | 0.0 | 9.40 | 3.0 | 0.0 | 'Less than 5 hours' | Healthy | MSc | No | 0.0 | 3.0 | Yes | 0 |
| 27898 | 140689 | Male | 31.0 | Faridabad | Student | 3.0 | 0.0 | 6.61 | 4.0 | 0.0 | '5-6 hours' | Unhealthy | MD | No | 12.0 | 2.0 | No | 0 |
| 27899 | 140690 | Female | 18.0 | Ludhiana | Student | 5.0 | 0.0 | 6.88 | 2.0 | 0.0 | 'Less than 5 hours' | Healthy | 'Class 12' | Yes | 10.0 | 5.0 | No | 1 |
| 27900 | 140699 | Male | 27.0 | Patna | Student | 4.0 | 0.0 | 9.24 | 1.0 | 0.0 | 'Less than 5 hours' | Healthy | BCA | Yes | 2.0 | 3.0 | Yes | 1 |
27901 rows × 18 columns
In [12]:
import pandas as pd
import matplotlib.pyplot as plt
grouped = data.groupby("Age")["Depression"].mean()
grouped.plot(kind = "bar")
plt.xlabel("Age")
plt.ylabel("Depression")
plt.title("Student Depression")
plt.show()
Functions and Fitting¶
Polyfit and Linear¶
In [13]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# ----------------------------------
# LOAD YOUR DATA
# ----------------------------------
data = pd.read_csv("~/work/rinchen-khandu/datasets/student_depression_dataset.csv")
# Select columns
x_col = "Age"
y_col = "Depression"
# Drop missing values
df = data[[x_col, y_col]].dropna()
x = df[x_col].values
y = df[y_col].values
np.set_printoptions(precision=3)
# ----------------------------------
# POLYNOMIAL FITTING
# ----------------------------------
# First-order (linear) fit
coeff1 = np.polyfit(x, y, 1)
pfit1 = np.poly1d(coeff1)
# Second-order (quadratic) fit
coeff2 = np.polyfit(x, y, 2)
pfit2 = np.poly1d(coeff2)
print(f"First-order fit coefficients (linear): {coeff1}")
print(f"Second-order fit coefficients (quadratic): {coeff2}")
# ----------------------------------
# CREATE SMOOTH FIT CURVES
# ----------------------------------
xmin, xmax = x.min(), x.max()
xfit = np.linspace(xmin, xmax, 200)
yfit1 = pfit1(xfit)
yfit2 = pfit2(xfit)
# ----------------------------------
# PLOTTING
# ----------------------------------
plt.figure(figsize=(8, 6))
plt.plot(x, y, 'o', alpha=0.6, label="Observed data")
plt.plot(xfit, yfit1, 'g-', linewidth=2, label="Linear fit")
plt.plot(xfit, yfit2, 'r-', linewidth=2, label="Quadratic fit")
plt.xlabel(x_col)
plt.ylabel(y_col)
plt.title("Linear vs Quadratic Fit using Real Data")
plt.legend()
plt.grid(True)
plt.show()
First-order fit coefficients (linear): [-0.023 1.173] Second-order fit coefficients (quadratic): [-0.001 0.019 0.651]
In [14]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# ----------------------------------
# LOAD YOUR DATA
# ----------------------------------
data = pd.read_csv("~/work/rinchen-khandu/datasets/student_depression_dataset.csv")
# Choose columns
x_col = "Age" # independent variable
y_col = "Depression" # dependent variable
# Drop missing values
df = data[[x_col, y_col]].dropna()
x = df[x_col].values
y = df[y_col].values
# ----------------------------------
# SORT DATA (IMPORTANT FOR SMOOTH PLOTS)
# ----------------------------------
idx = np.argsort(x)
x = x[idx]
y = y[idx]
xmin, xmax = x.min(), x.max()
npts = len(x)
# ----------------------------------
# FIT POLYNOMIALS OF DIFFERENT ORDERS
# ----------------------------------
xplot = np.linspace(xmin - 0.2, xmax + 0.2, 300)
# Order 1 (linear)
coeff1 = np.polyfit(x, y, 1)
yfit1 = np.poly1d(coeff1)(xplot)
# Order 4 (moderate complexity)
coeff4 = np.polyfit(x, y, 4)
yfit4 = np.poly1d(coeff4)(xplot)
# Order 15 (high-degree / overfitting)
coeff15 = np.polyfit(x, y, 15)
yfit15 = np.poly1d(coeff15)(xplot)
# ----------------------------------
# PLOTTING
# ----------------------------------
fig = plt.figure(figsize=(8, 6))
fig.canvas.header_visible = False
plt.plot(x, y, 'bo', alpha=0.6, label='Observed data')
plt.plot(xplot, yfit1, 'g-', linewidth=2, label='Order 1 (Underfit)')
plt.plot(xplot, yfit4, 'c-', linewidth=2, label='Order 4')
plt.plot(xplot, yfit15, 'r-', linewidth=2, label='Order 15 (Overfit)')
plt.xlabel(x_col)
plt.ylabel(y_col)
plt.title("Polynomial Fit Comparison Using Real Data")
plt.legend()
plt.grid(True)
plt.show()
/tmp/ipykernel_26369/1382633408.py:44: RankWarning: Polyfit may be poorly conditioned coeff15 = np.polyfit(x, y, 15)
Probability¶
In [17]:
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
#
# load your own data
#
data = pd.read_csv("~/work/rinchen-khandu/datasets/student_depression_dataset.csv")
# ✅ choose the column to analyze
x = data["Age"].dropna().values # change to "Age" if needed
npts = len(x)
#
# estimate Gaussian parameters from your data
#
mean = np.mean(x)
stddev = np.std(x)
#
# plot histogram and data points
#
plt.hist(x, bins=npts // 50, density=True, alpha=0.6)
plt.plot(x, np.zeros_like(x), '|', ms=10)
#
# plot fitted Gaussian curve
#
xi = np.linspace(mean - 3 * stddev, mean + 3 * stddev, 200)
yi = np.exp(-(xi - mean) ** 2 / (2 * stddev ** 2)) / np.sqrt(2 * np.pi * stddev ** 2)
plt.plot(xi, yi, 'r', linewidth=2)
plt.xlabel("Depression Score")
plt.ylabel("Probability Density")
plt.title("Gaussian Fit to Student Depression Data")
plt.show()
Density and Estimation¶
Voronoi tesselation¶
In [18]:
import matplotlib.pyplot as plt
from scipy.spatial import Voronoi, voronoi_plot_2d
import numpy as np
import pandas as pd
#
# k-means parameters
#
nsteps = 1000
momentum = 0. # (kept for consistency, not used)
#
# import data from CSV
#
data = pd.read_csv('~/work/rinchen-khandu/datasets/student_depression_dataset.csv')
# ✅ FIX: Use Age and Depression columns
# Drop missing values to avoid errors
data = data[["Age", "Depression"]].dropna()
x = data["Age"].values
y = data["Depression"].values
#
# k-means function
#
def kmeans(x, y, momentum, nclusters):
# choose random initial cluster centers
indices = np.random.randint(0, len(x), nclusters)
mux = x[indices]
muy = y[indices]
for _ in range(nsteps):
# distance of each point to each cluster center
X = np.outer(x, np.ones(len(mux)))
Y = np.outer(y, np.ones(len(muy)))
Mux = np.outer(np.ones(len(x)), mux)
Muy = np.outer(np.ones(len(x)), muy)
distances = np.sqrt((X - Mux)**2 + (Y - Muy)**2)
mins = np.argmin(distances, axis=1)
# update cluster centers
for i in range(len(mux)):
index = np.where(mins == i)[0]
if len(index) > 0:
mux[i] = np.mean(x[index])
muy[i] = np.mean(y[index])
# compute total distance (for elbow method)
total_distance = 0
for i in range(len(mux)):
index = np.where(mins == i)[0]
total_distance += np.sum(
np.sqrt((x[index] - mux[i])**2 + (y[index] - muy[i])**2)
)
return mux, muy, total_distance
#
# plot k-means clusters
#
def plot_kmeans(x, y, mux, muy):
plt.figure()
plt.plot(x, y, '.', alpha=0.6)
plt.plot(mux, muy, 'r.', markersize=20)
plt.xlabel("Age")
plt.ylabel("Depression Score")
plt.title(f"{len(mux)} clusters (K-Means)")
plt.show()
#
# plot Voronoi diagram
#
def plot_Voronoi(x, y, mux, muy):
plt.figure()
plt.plot(x, y, '.', alpha=0.6)
vor = Voronoi(np.column_stack((mux, muy)))
voronoi_plot_2d(
vor,
show_points=True,
show_vertices=False,
point_size=20
)
plt.xlabel("Age")
plt.ylabel("Sleep Duration")
plt.title(f"{len(mux)} clusters (Voronoi)")
plt.show()
#
# run clustering for different cluster counts
#
distances = np.zeros(5)
mux, muy, distances[0] = kmeans(x, y, momentum, 1)
plot_kmeans(x, y, mux, muy)
mux, muy, distances[1] = kmeans(x, y, momentum, 2)
plot_kmeans(x, y, mux, muy)
mux, muy, distances[2] = kmeans(x, y, momentum, 3)
plot_Voronoi(x, y, mux, muy)
mux, muy, distances[3] = kmeans(x, y, momentum, 4)
plot_Voronoi(x, y, mux, muy)
mux, muy, distances[4] = kmeans(x, y, momentum, 5)
plot_Voronoi(x, y, mux, muy)
#
# elbow method plot
#
plt.figure()
plt.plot(np.arange(1, 6), distances, 'o-')
plt.xlabel("Number of clusters")
plt.ylabel("Total distance to clusters")
plt.title("Elbow Method (Age vs Depression)")
plt.show()
--------------------------------------------------------------------------- QhullError Traceback (most recent call last) Cell In[18], line 109 106 plot_kmeans(x, y, mux, muy) 108 mux, muy, distances[2] = kmeans(x, y, momentum, 3) --> 109 plot_Voronoi(x, y, mux, muy) 111 mux, muy, distances[3] = kmeans(x, y, momentum, 4) 112 plot_Voronoi(x, y, mux, muy) Cell In[18], line 83, in plot_Voronoi(x, y, mux, muy) 80 plt.figure() 81 plt.plot(x, y, '.', alpha=0.6) ---> 83 vor = Voronoi(np.column_stack((mux, muy))) 84 voronoi_plot_2d( 85 vor, 86 show_points=True, 87 show_vertices=False, 88 point_size=20 89 ) 91 plt.xlabel("Age") File scipy/spatial/_qhull.pyx:2680, in scipy.spatial._qhull.Voronoi.__init__() File scipy/spatial/_qhull.pyx:356, in scipy.spatial._qhull._Qhull.__init__() QhullError: QH6154 Qhull precision error: Initial simplex is flat (facet 1 is coplanar with the interior point) While executing: | qhull v Qc Qz Qbb Options selected for Qhull 2020.2.r 2020/08/31: run-id 1695218816 voronoi Qcoplanar-keep Qz-infinity-point Qbbound-last _pre-merge _zero-centrum Qinterior-keep Pgood _max-width 11 Error-roundoff 4.3e-14 _one-merge 3e-13 Visible-distance 8.6e-14 U-max-coplanar 8.6e-14 Width-outside 1.7e-13 _wide-facet 5.1e-13 _maxoutside 3.4e-13 The input to qhull appears to be less than 3 dimensional, or a computation has overflowed. Qhull could not construct a clearly convex simplex from points: - p3(v4): 25 0 31 - p0(v3): 25 0 10 - p2(v2): 31 0 26 - p1(v1): 20 0 0 The center point is coplanar with a facet, or a vertex is coplanar with a neighboring facet. The maximum round off error for computing distances is 4.3e-14. The center point, facets and distances to the center point are as follows: center point 25.2 0 16.91 facet p0 p2 p1 distance= 0 facet p3 p2 p1 distance= 0 facet p3 p0 p1 distance= 0 facet p3 p0 p2 distance= 0 These points either have a maximum or minimum x-coordinate, or they maximize the determinant for k coordinates. Trial points are first selected from points that maximize a coordinate. The min and max coordinates for each dimension are: 0: 19.92 30.87 difference= 10.95 1: 0 0 difference= 0 2: 0 30.87 difference= 30.87 If the input should be full dimensional, you have several options that may determine an initial simplex: - use 'QJ' to joggle the input and make it full dimensional - use 'QbB' to scale the points to the unit cube - use 'QR0' to randomly rotate the input for different maximum points - use 'Qs' to search all points for the initial simplex - use 'En' to specify a maximum roundoff error less than 4.3e-14. - trace execution with 'T3' to see the determinant for each point. If the input is lower dimensional: - use 'QJ' to joggle the input and make it full dimensional - use 'Qbk:0Bk:0' to delete coordinate k from the input. You should pick the coordinate with the least range. The hull will have the correct topology. - determine the flat containing the points, rotate the points into a coordinate plane, and delete the other coordinates. - add one or more points to make the input full dimensional.
Modeling Functions¶
In [19]:
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
#
# Load your own data
#
data = pd.read_csv("~/work/rinchen-khandu/datasets/student_depression_dataset.csv")
data = data[["Age", "Depression"]].dropna()
x = data["Age"].values
y = data["Depression"].values
# sort data (important for plotting smooth curves)
idx = np.argsort(x)
x = x[idx]
y = y[idx]
npts = len(x)
#
# function cluster-weighted modeling parameters
#
nclusters = 5
minvar = 0.01
niterations = 100
np.random.seed(10)
xplot = np.copy(x)
nplot = npts
#
# initialize parameters
#
mux = np.random.choice(x, nclusters)
beta = 0.1 * np.random.rand(3, nclusters)
varx = np.ones(nclusters)
vary = np.ones(nclusters)
pc = np.ones(nclusters) / nclusters
#
# ---------- BEFORE ITERATION ----------
#
pxgc = np.exp(-(np.outer(x, np.ones(nclusters))
- np.outer(np.ones(npts), mux))**2
/ (2 * np.outer(np.ones(npts), varx))) \
/ np.sqrt(2 * np.pi * np.outer(np.ones(npts), varx))
pygxc = np.exp(-(np.outer(y, np.ones(nclusters))
- (np.outer(np.ones(npts), beta[0, :])
+ np.outer(x, beta[1, :])
+ np.outer(x * x, beta[2, :])))**2
/ (2 * np.outer(np.ones(npts), vary))) \
/ np.sqrt(2 * np.pi * np.outer(np.ones(npts), vary))
pxc = pxgc * np.outer(np.ones(npts), pc)
pxyc = pygxc * pxc
pcgxy = pxyc / np.outer(np.sum(pxyc, 1), np.ones(nclusters))
plt.figure()
plt.plot(x, y, '.', alpha=0.6)
yplot = np.sum((np.outer(np.ones(nplot), beta[0, :])
+ np.outer(x, beta[1, :])
+ np.outer(x * x, beta[2, :])) * pxc, 1) / np.sum(pxc, 1)
plt.plot(xplot, yplot)
yvarplot = np.sum((vary +
(np.outer(np.ones(nplot), beta[0, :])
+ np.outer(x, beta[1, :])
+ np.outer(x * x, beta[2, :]))**2) * pxc, 1) \
/ np.sum(pxc, 1) - yplot**2
ystdplot = np.sqrt(yvarplot)
plt.plot(xplot, yplot + ystdplot, 'k--')
plt.plot(xplot, yplot - ystdplot, 'k--')
plt.title("Function Clusters BEFORE EM Iteration")
plt.xlabel("Age")
plt.ylabel("Depression Score")
plt.show()
#
# ---------- EM ITERATION ----------
#
for step in range(niterations):
pxgc = np.exp(-(np.outer(x, np.ones(nclusters))
- np.outer(np.ones(npts), mux))**2
/ (2 * np.outer(np.ones(npts), varx))) \
/ np.sqrt(2 * np.pi * np.outer(np.ones(npts), varx))
pygxc = np.exp(-(np.outer(y, np.ones(nclusters))
- (np.outer(np.ones(npts), beta[0, :])
+ np.outer(x, beta[1, :])
+ np.outer(x * x, beta[2, :])))**2
/ (2 * np.outer(np.ones(npts), vary))) \
/ np.sqrt(2 * np.pi * np.outer(np.ones(npts), vary))
pxc = pxgc * np.outer(np.ones(npts), pc)
pxyc = pygxc * pxc
pcgxy = pxyc / np.outer(np.sum(pxyc, 1), np.ones(nclusters))
pc = np.sum(pcgxy, 0) / npts
mux = np.sum(np.outer(x, np.ones(nclusters)) * pcgxy, 0) / (npts * pc)
varx = minvar + np.sum((np.outer(x, np.ones(nclusters))
- np.outer(np.ones(npts), mux))**2
* pcgxy, 0) / (npts * pc)
a = np.array([
np.sum(np.outer(y, np.ones(nclusters)) * pcgxy, 0) / (npts * pc),
np.sum(np.outer(y * x, np.ones(nclusters)) * pcgxy, 0) / (npts * pc),
np.sum(np.outer(y * x * x, np.ones(nclusters)) * pcgxy, 0) / (npts * pc)
])
B = np.zeros((3, 3, nclusters))
for c in range(nclusters):
B[:, :, c] = [
[1,
np.sum(x * pcgxy[:, c]) / (npts * pc[c]),
np.sum(x * x * pcgxy[:, c]) / (npts * pc[c])],
[np.sum(x * pcgxy[:, c]) / (npts * pc[c]),
np.sum(x * x * pcgxy[:, c]) / (npts * pc[c]),
np.sum(x * x * x * pcgxy[:, c]) / (npts * pc[c])],
[np.sum(x * x * pcgxy[:, c]) / (npts * pc[c]),
np.sum(x * x * x * pcgxy[:, c]) / (npts * pc[c]),
np.sum(x * x * x * x * pcgxy[:, c]) / (npts * pc[c])]
]
beta[:, c] = np.linalg.inv(B[:, :, c]) @ a[:, c]
vary = minvar + np.sum((np.outer(y, np.ones(nclusters))
- (np.outer(np.ones(npts), beta[0, :])
+ np.outer(x, beta[1, :])
+ np.outer(x * x, beta[2, :])))**2
* pcgxy, 0) / (npts * pc)
#
# ---------- AFTER ITERATION ----------
#
plt.figure()
plt.plot(x, y, '.', alpha=0.6)
yplot = np.sum((np.outer(np.ones(nplot), beta[0, :])
+ np.outer(x, beta[1, :])
+ np.outer(x * x, beta[2, :])) * pxc, 1) / np.sum(pxc, 1)
plt.plot(xplot, yplot)
ystdplot = np.sqrt(yvarplot)
plt.plot(xplot, yplot + ystdplot, 'k--')
plt.plot(xplot, yplot - ystdplot, 'k--')
plt.title("Function Clusters AFTER EM Iteration")
plt.xlabel("Age")
plt.ylabel("Depression Score")
plt.show()
/tmp/ipykernel_26369/3832536625.py:58: RuntimeWarning: invalid value encountered in divide pcgxy = pxyc / np.outer(np.sum(pxyc, 1), np.ones(nclusters))
/tmp/ipykernel_26369/3832536625.py:102: RuntimeWarning: invalid value encountered in divide pcgxy = pxyc / np.outer(np.sum(pxyc, 1), np.ones(nclusters))
Transformation¶
In [20]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
np.set_printoptions(precision=2)
#
# load your own data
#
data = pd.read_csv("~/work/rinchen-khandu/datasets/student_depression_dataset.csv")
# select numerical columns only
numerical_cols = data.select_dtypes(include=[np.number]).columns
X = data[numerical_cols].dropna().values
print(f"Data shape (records, features): {X.shape}")
#
# optional: plot vs first two columns
#
plt.scatter(X[:, 0], X[:, 1], c='blue', alpha=0.6)
plt.xlabel(numerical_cols[0])
plt.ylabel(numerical_cols[1])
plt.title(f"{numerical_cols[0]} vs {numerical_cols[1]}")
plt.show()
#
# standardize data (zero mean, unit variance)
#
X_mean = np.mean(X, axis=0)
X_std = np.std(X, axis=0)
Xscale = (X - X_mean) / np.where(X_std > 0, X_std, 1)
print(f"Standardized data mean: {np.mean(Xscale):.2f}, variance: {np.var(Xscale):.2f}")
#
# do PCA
#
n_components = min(50, Xshape := Xscale.shape[1]) # max 50 or # of features
pca = PCA(n_components=n_components)
pca.fit(Xscale)
Xpca = pca.transform(Xscale)
#
# plot explained variance
#
plt.plot(pca.explained_variance_, 'o-')
plt.xlabel('PCA component')
plt.ylabel('Explained variance')
plt.title('PCA Explained Variance')
plt.show()
#
# plot first two PCA components
#
plt.scatter(Xpca[:, 0], Xpca[:, 1], c='green', s=20, alpha=0.6)
plt.xlabel("Principal component 1")
plt.ylabel("Principal component 2")
plt.title("Data projected on first two PCA components")
plt.show()
Data shape (records, features): (27901, 9)
Standardized data mean: 0.00, variance: 1.00
References¶
- ChatGPT
- Class notes
- Help sessions
- Support from friends
We would like to thank Data Science Team, Professor Niel Gershenfeld and Team for supporting us with Data Science Sessions.
Thank You